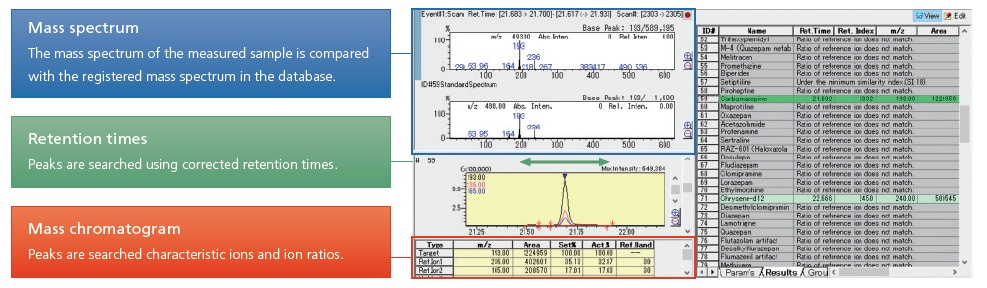
Base de données toxicologiques médico-légales GC/MS - Fonctionnalités
Pour l'analyse toxicologique GC/MS
Simultaneous toxicology screening with a high-sensitivity scanning method
The GC/MS scanning method is the gold-standard approach to toxicology analysis, indispensable for the identification of toxicological substances.
Data processing using the mass spectrum library involves looking up the detected peaks and judging whether they correspond to a known drug compound. However, target peaks may be missed if they overlap with interference peaks from the biological sample, or if they are buried in a TIC (total ion current chromatogram) because of their low concentration.
With this database, the mass chromatogram is used to search for peaks and identify them based on three types of information stored in the database: retention time, ion ratio and mass spectrum. In this way, the presence or absence of each drug can be verified, and there is no need to check the TIC, which often contains a large number of impurities. Data processing can be carried out more quickly while minimizing the risk of overlooking any target compounds.
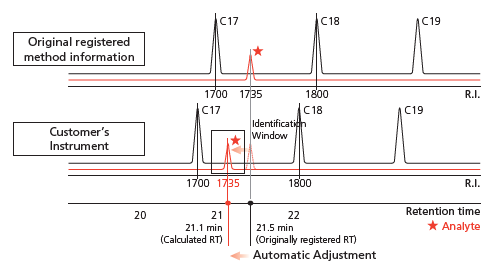
Precisely correct the retention times of all listed compounds using AART
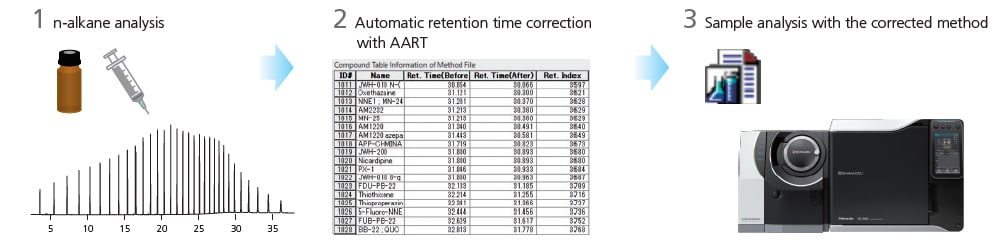
The retention times of all compounds listed in the method file can be simultaneously corrected to your GC-MS system with the Automatic Adjustment of Retention Time (AART) function. The multiple n-alkanes used as references from low to high boiling points allow precise correction over a wide range of retention times.
Support for volatile toxic substances with an HS-20 headspace sampler
The database provides optimum analysis conditions for blood alcohol and volatile toxic substances, indispensable for forensic police laboratories and academic forensics. Volatile toxic substances in the database include cyanides, azides, and thinners such as methanol, ethyl acetate, and toluene.
The database also includes a method for calculating semi-quantitative values. Simply add internal standards to the test sample before measurement to estimate its concentration. This is useful for setting the concentration range of the calibration curve for precise quantitation.
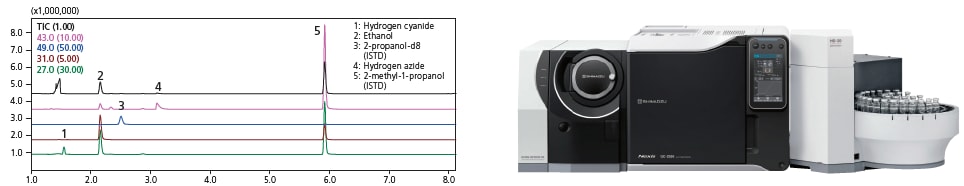
Simultaneous analysis of blood alcohol and volatile toxic substances using an HS-20 headspace sampler
Comprehensive detection of designer drugs with MS/MS mode
The MS/MS mode of the GCMS-TQ series enables the user to comprehensively detect and estimate the likely structure of cathinones or synthetic naphthoylindole cannabinoids.
■Comprehensive detection of cathinones
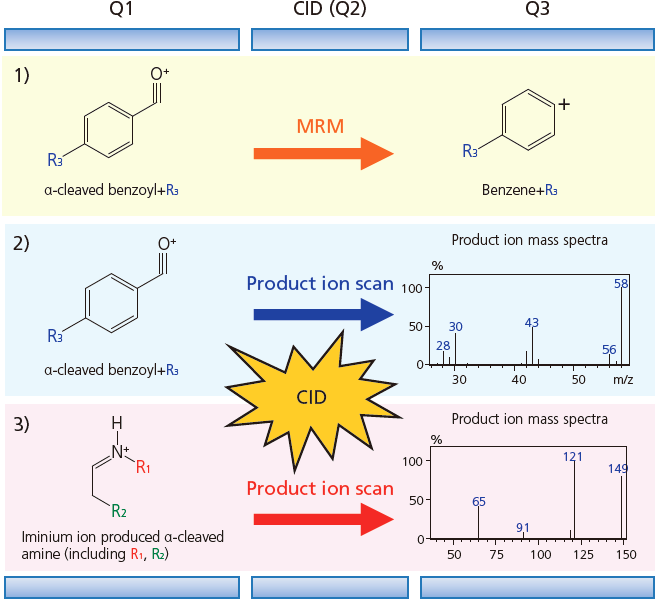
It is possible to comprehensively detect approx. 1300 types of cathinone via α-cleavage into benzoyl and amine ions which are measured by MRM and product ion scans with GC-MS/MS. Cathinone structure can be estimated from the MS/MS results. It is difficult to distinguish the functional group on the amine with an EI scan due to the large number of isomers, but it is relatively simple to differentiate structural isomers with a product ion scan.
1) The cathinone structure is identified and the R3 functional group estimated
2) The R3 functional group is estimated and the positional isomer distinguished
3) The R1 and R2 functional groups are estimated and the structural isomer distinguished
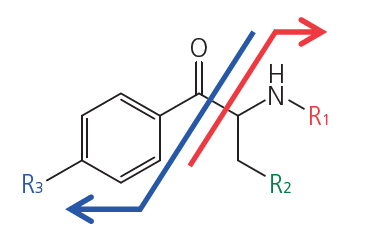
Cathinone structure
■Comprehensive detection of synthetic naphthoylindole cannabinoids
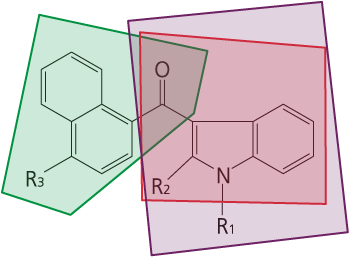
Basic structure of a naphthoylindole cannabinoid
It is possible to comprehensively detect approx. 800 kinds of naphthoylindole cannabinoids via EI ionization cleavage into a naphthoylindole and an indole carbonyl. Structure of these parts can be inferred from GC-MS/MS precursor ion scans and MRM.
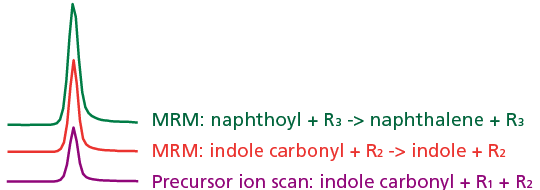
The three peaks from the different MS/MS modes have the same retention times, so it is possible to comprehensively detect all naphthoylindoles and estimate the functional groups R1-R3.


